
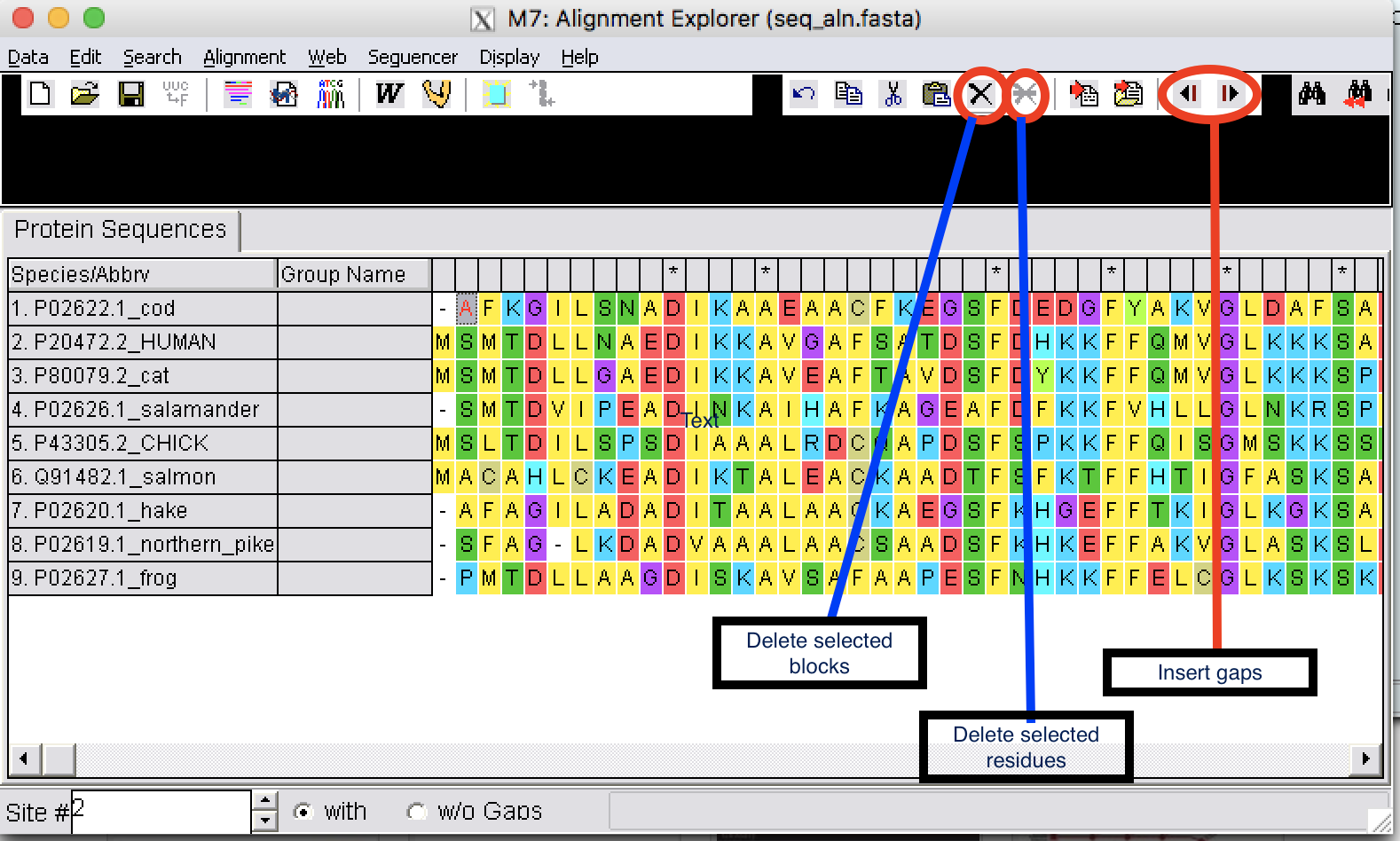
- #How to convert a file to fasta format in mega 7 software
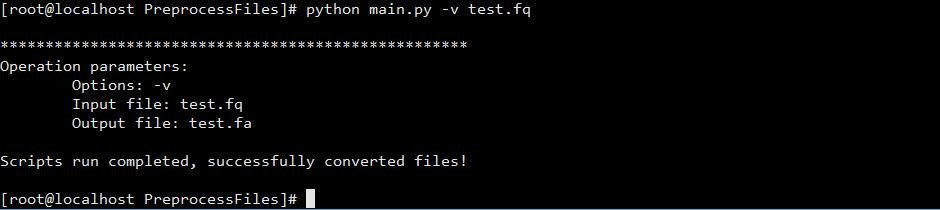
- #How to convert a file to fasta format in mega 7 code
Digits or else are removed and ignored (but Tab and space characters with combination end line character (Enter press) can be interpreted as column format). It read only standard IUB/IUPAC amino acid or nucleic acid codes characters and rejects anything else, low- and upper-case insensitive. Like a text/plain format without white space and TABs. You can modify a nucleotide sequences by inserting, deleting and replacing sequence fragments. When sequences are imported you may edit the sequences in general or additional editors and immediately visualize the result of editing. Sequence formats are simply the way in which the amino acid or DNA sequence is recorded in a computer file.
#How to convert a file to fasta format in mega 7 code
Importing sequences must be at the same format.ĭegenerate DNA sequences are accepted as IUPAC code is an extended vocabulary of 11 letters which allows the description of ambiguous DNA code.Įach letter represents a combination of one or several nucleotides: M=(A/C) R=(A/G) W=(A/T) S=(G/C) Y=(C/T) K=(G/T) V=(A/G/C) H=(A/C/T) D=(A/G/T) B=(C/G/T) N=(A/G/C/T), U=T and I (Inosine).Īccepted amino acid codes: A(Ala), C(Cys), D(Asp), E(Glu), F(Phe), G(Cly), H(His), I(Ile), K(Lys), L(Leu), M(Met), N(Asn), P(Pro), Q(Gln), R(Arg), S(Ser), T(Thr), U(Sec), V(Val), W(Trp), Y(Tyr). Import sequences are not different for two editors: both editors take all ways, with keyboard ( Shit-Insert, Ctrl-V) and allowed work with right-click mouse displays a contextual menu.
#How to convert a file to fasta format in mega 7 software
Software opened any text file with command line: fastpcr.exe filename.fastaįastPCR normally expect to read sequence files in FASTA format. Each file will converted to FASTA format with name of the file. Especially this is reliable for read all sequences from different files. You need click on any file from folder and will see result.
Files can be saved various formats including. The information status bar shows the number of sequences, the total sequence length (in nucleotides), the nucleotide composition and the purine, pyrimidine, CG percentage and the melting temperature. When a sequence file is opened, FastPCR displays the information about the opened sequence and its format. Alternatively this feature allows splitting FASTA sequences to individual files in a particular selected folder. For example, this feature can be applied to convert all files from a selected folder into a single file of FASTA sequences. Additionally, the programme can open files within a selected folder in order to join all these files in a text editor. Users can type or import from file(s) into "General Sequence(s)" or "Additional sequence(s) or pre-designed primers (probes) list" editors.įastPCR allows files to opened in several ways: the original file can opened as read-only for editing with text editors files can be opened to memory without using text editors, which allows larger file(s), up to 200Mb, to be analysed files within a folder can be selected and the files opened during task execution without the use of text editor programme. It is important that all target sequences are prepared in the same format. The FastPCR clipboard allows the user to copy and paste text or tables from MS Word documents or MS Excel worksheets or other programmes and to paste them into another Office document. The programme takes either a single sequence or accepts multiple separate DNA sequences in FASTA, tabulated format (two columns from MS Excel sheet or MS Word table), EMBL, MEGA, GenBank, MSF, DIALIGN, simple alignment, or BLAST Queue web alignment formats. Sequence data files are prepared using a text editor (Notepad, WordPad, MS Word), and saved in ASCII as text/plain format (.


 0 kommentar(er)
0 kommentar(er)
